Harmony
What is Harmony?
Harmony is a centralized online workspace for tabular biological data management and analysis. Use Harmony to organize studies into projects, data and metadata, invite contributors, and perform standardized analyses.
Why go with Harmony?
To enable enterprise-wide visibility into past and ongoing experiments, clinical studies, and relevant public data—all just a few clicks away from reanalysis as new hypotheses emerge. Harmony is the ultimate FAIR platform.
Harmony simplified
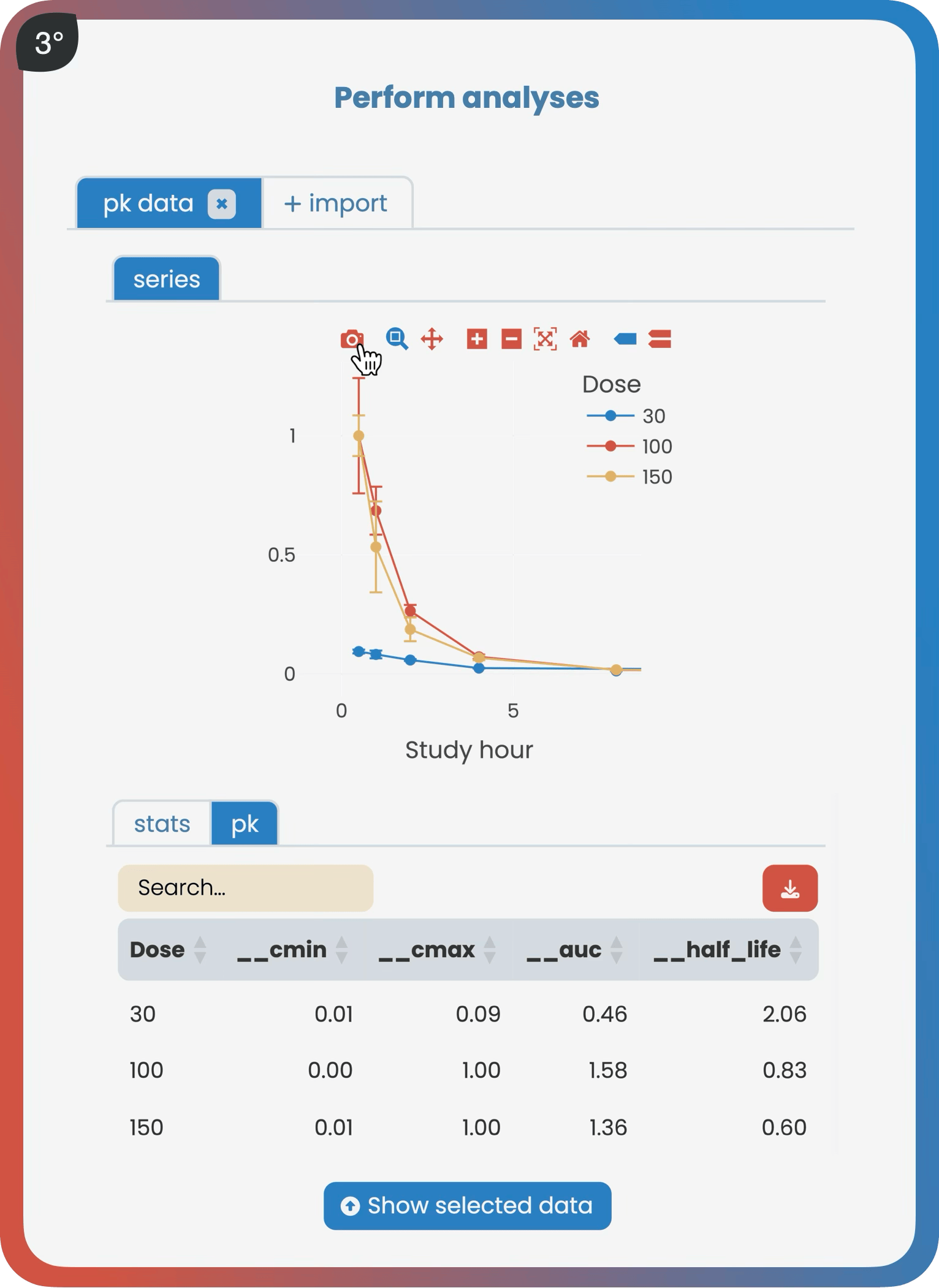
➔ Swipe through a typical workflow ➔
What you’ll get
A centralized online workspace
Use SSO credentials to access Bioinframed’s online infrastructure.
Trainings & support
Attend trainings and receive hands-on data entry assistance.
Granular access control
Control access permissions down to the dataset level.
Guided, intuitive data entry
Add & edit data in Harmony’s online spreadsheet-style interface that guides best-practice compliance.
A project-based database
Track current & past experiments, review their key results, and (re)analyze anytime.
Interactive analyses
Perform interactive, reproducible analyses on any object-over-variable relationship, single-cell data, spatial data, and more.
API connectivity
Seamlessly connect coding and non-coding scientists to collaboratively build a gold-standard database.
An ultra-fast architecture
Experience blazing performance thanks to cutting-edge libraries for in-memory operations and rapid I/O.
Harmony lets you manage & analyze
Manage
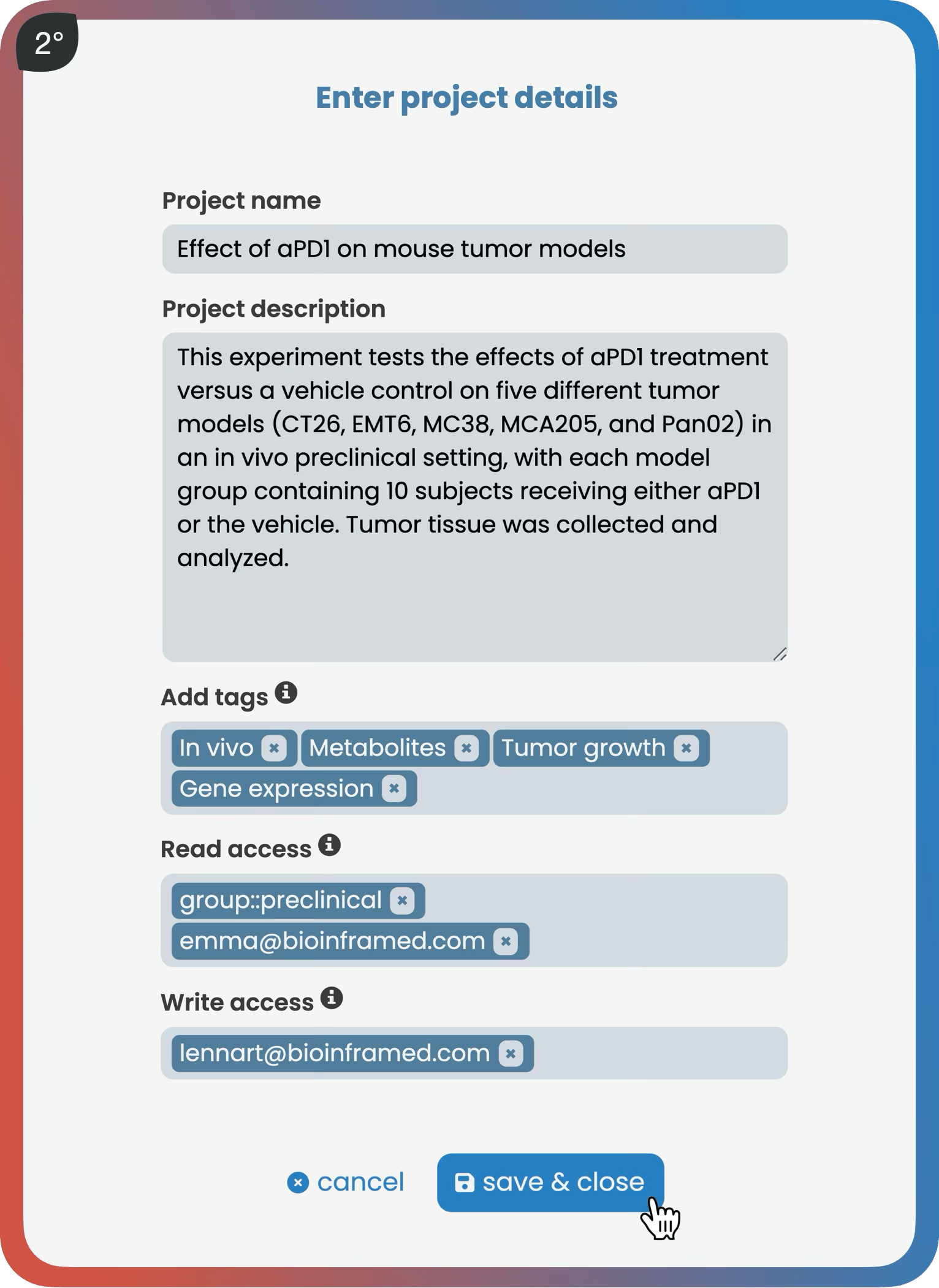
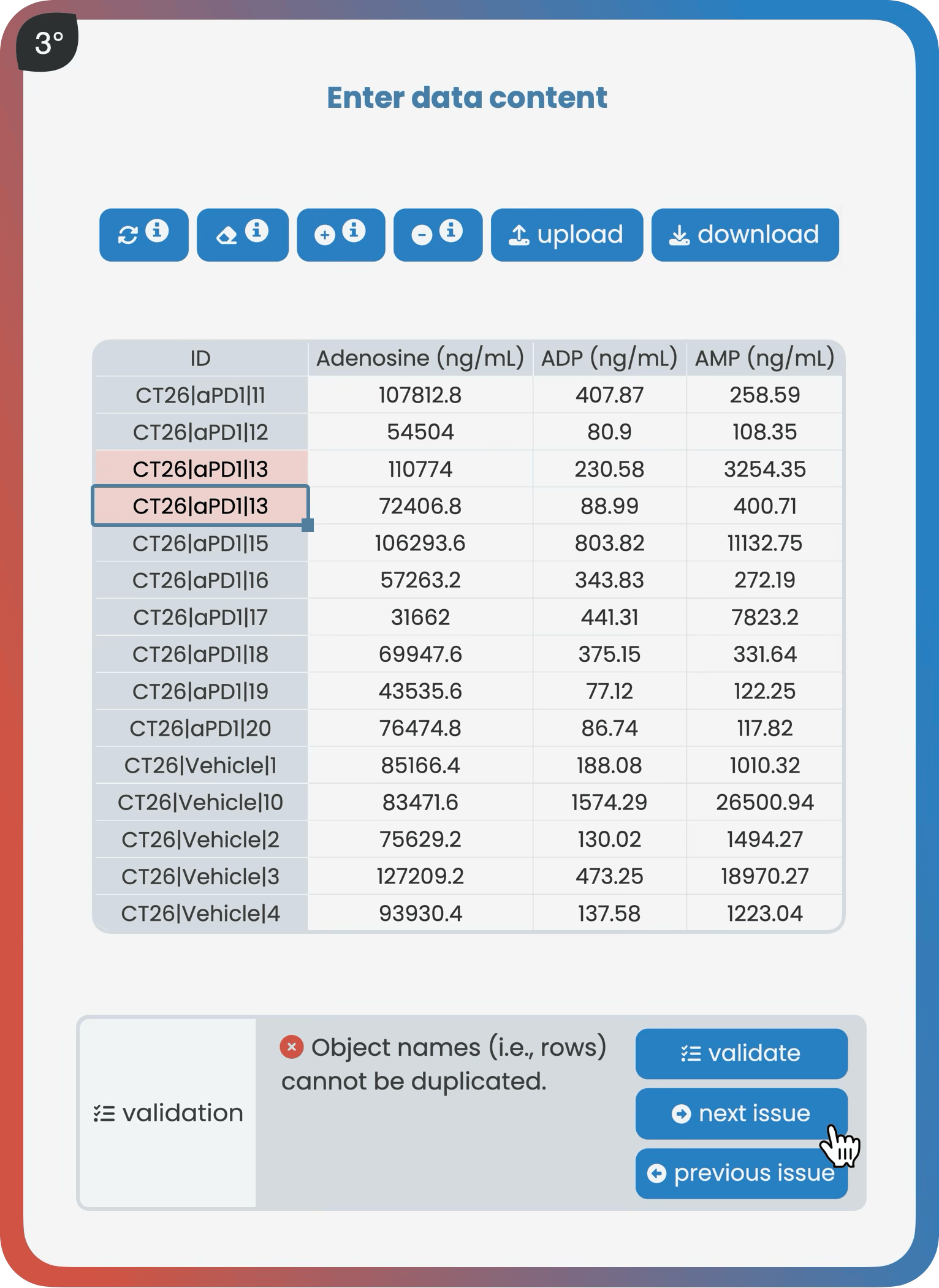
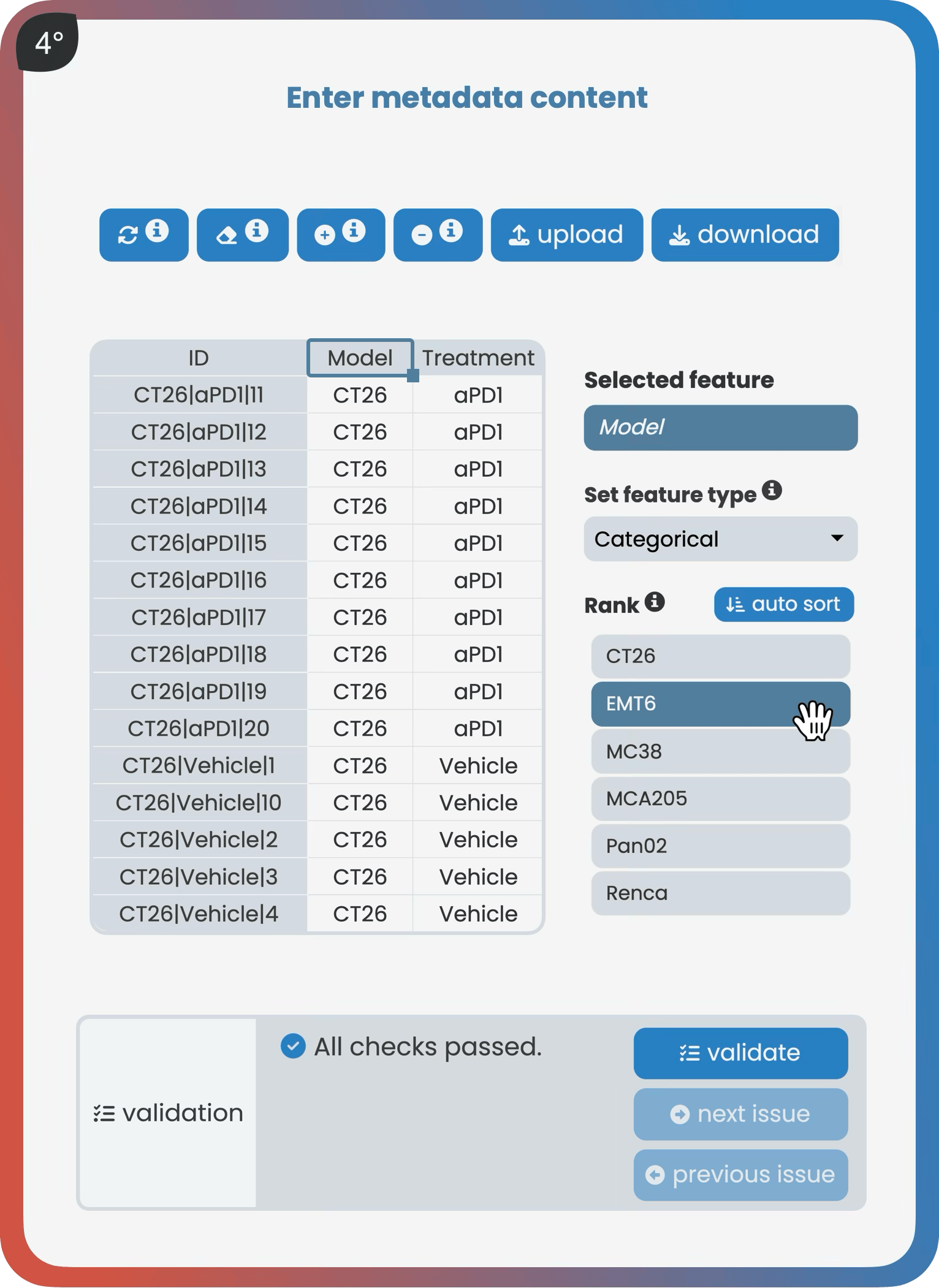
The manage page allows users to create projects, manage versions, add data and metadata, and set user access rights. As a core part of robust data management, it guides users to store their data following best practice guidelines—either through an interactive spreadsheet editor or via Harmony’s R API. Once datasets meet predefined standards, they receive a validation checkmark indicating they are ready for analysis within Harmony.
Analyze
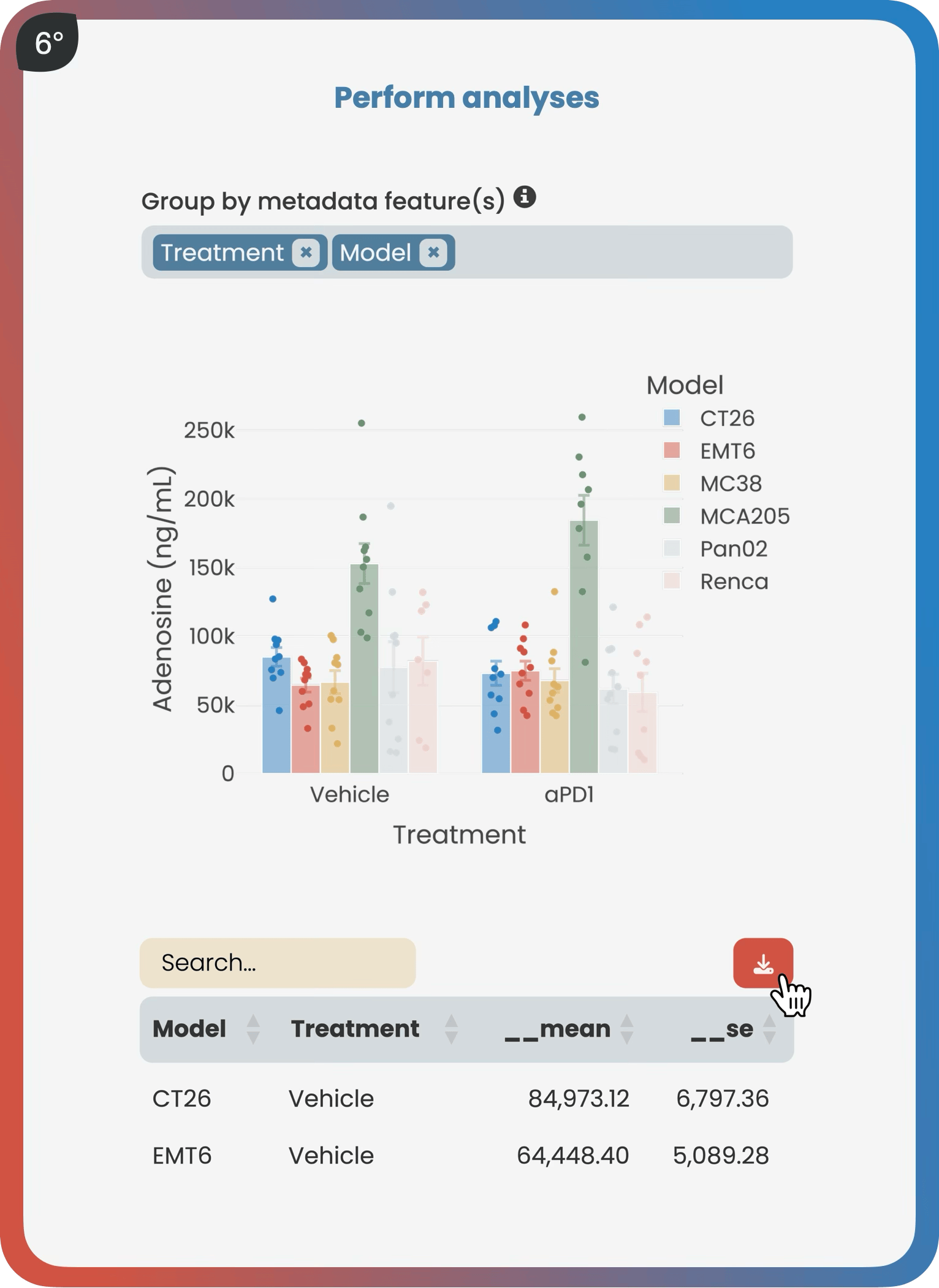
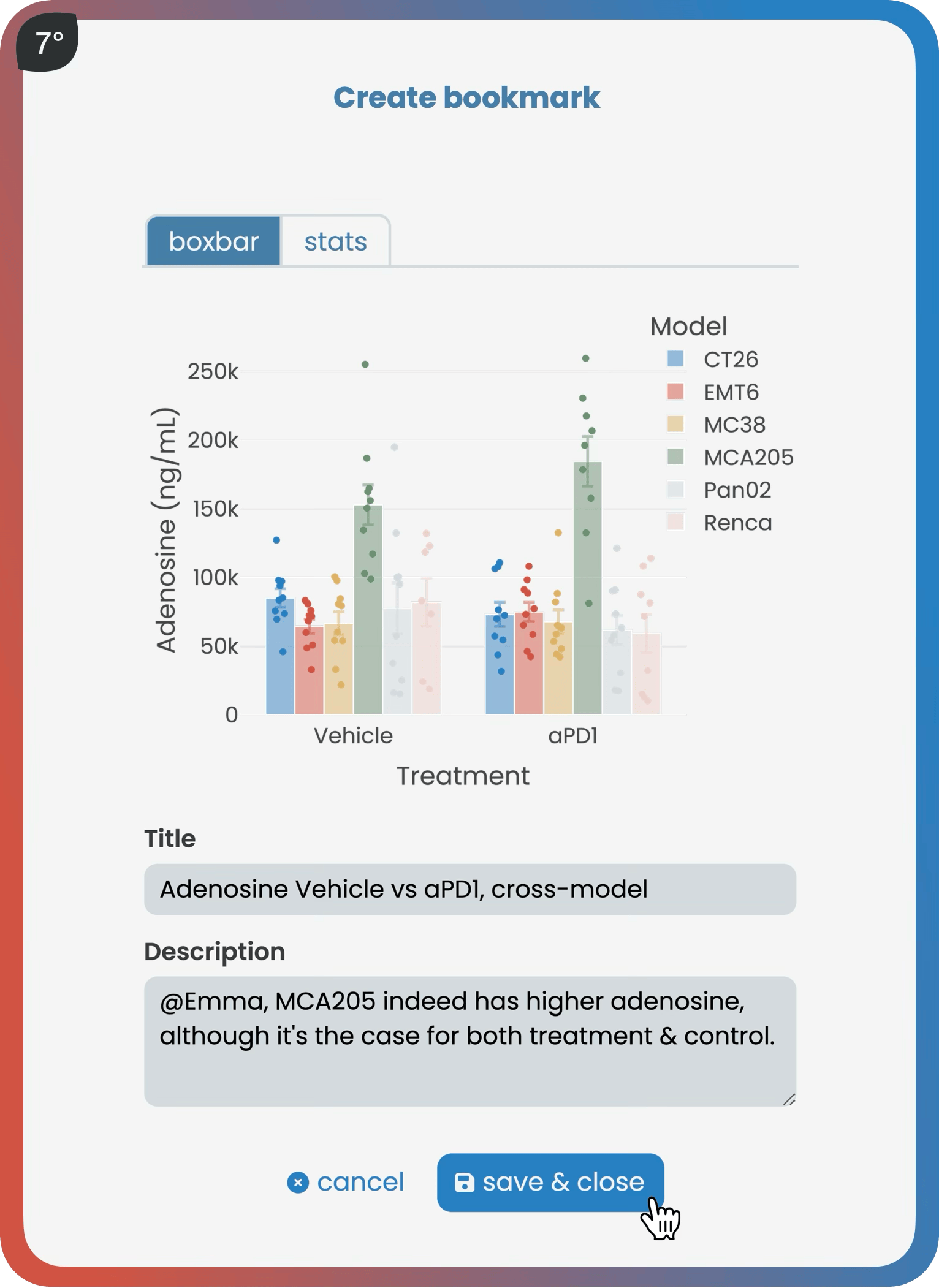
The analyze page provides interactive analysis tools applicable to successfully validated data. According to the type of data, like bulk RNA sequencing, users gain access to a suite of standardized analyses, such as a differential gene expression analysis. Bookmarking plots makes it easy for all project contributors to seamlessly share results.
View the full demo library here.
FAQ
What data types are currently supported?
-
The object-over-variable class fits most data types, where rows represent objects and columns represent variables. For example, a metabolomics dataset could have each row as a tissue sample (object) and each column as a metabolite such as adenosine (variable), with the entries holding the measured values.
Available analyses: Box & bar plots with statistical testing, time series, heatmaps, principal component analysis, correlation analysis, linear modeling, and more.
-
The survival class stores subject-level time points and events.
Available analyses: Kaplan–Meier curves, survival modeling, and hazard-based statistics.
-
The bulk RNA sequencing class is a specialization of the object-over-variable type, with raw read counts per gene as measured values.
Available analyses: see object-over-variable + differential expression, pathway over-representation, and gene set variation analysis.
-
The single-cell class stores high-dimensional single-cell data (e.g., scRNAseq), directly importable through Seurat, among others.
Available analyses: cluster analysis, box & bar plots, dot plots.
-
The spatial class extends the object-over-variable format by adding spatial coordinates, describing objects (cells or spots) not only by variables but also by their physical position.
Available analyses: cluster analysis, tissue-value mapping, box & bar plots, dot plots.
-
The blob class is storage-focused, optimized for (binary) files such as images, raw instrument outputs, or other unstructured datasets.
What challenges does Harmony address?
-
Researchers often visualize data independently using tools like Microsoft Excel, Tableau or GraphPad Prism, which inevitably requires a data cleaning (i.e., parsing) step. Similarly, bioinformaticians frequently spend time parsing and re-parsing data before analysis. Harmony eliminates this redundancy by enabling a one-time, collaborative data cleaning process, accessible to both coders and non-coders. Whether data is generated in-house, obtained from a sequencing provider, or delivered through a CRO data portal, this process creates a gold-standard, project-based database that everyone can trust and build upon.
-
Artificial intelligence is powerful, yet its success relies on something too often overlooked—clean interpretable data. Parsing and restructuring data can be cumbersome, but Harmony intuitively guides users through the process, with its extensive analytics framework providing a strong incentive towards best-practice data storage.
-
Harmony improves visibility into both ongoing and completed experiments, on top enabling quick access to key findings via its bookmarking features. Users can execute both general and field-specific queries across the entire database.
-
Harmony’s analytics framework offers a rapid standardized approach to efficient data analysis, covering a wide range of common tasks including box and bar plots with statistical testing, time series analysis, principal component analysis, pie charts, heatmaps, linear modeling, differential expression analysis, single-cell cluster analysis, spatial data exploration, and more.
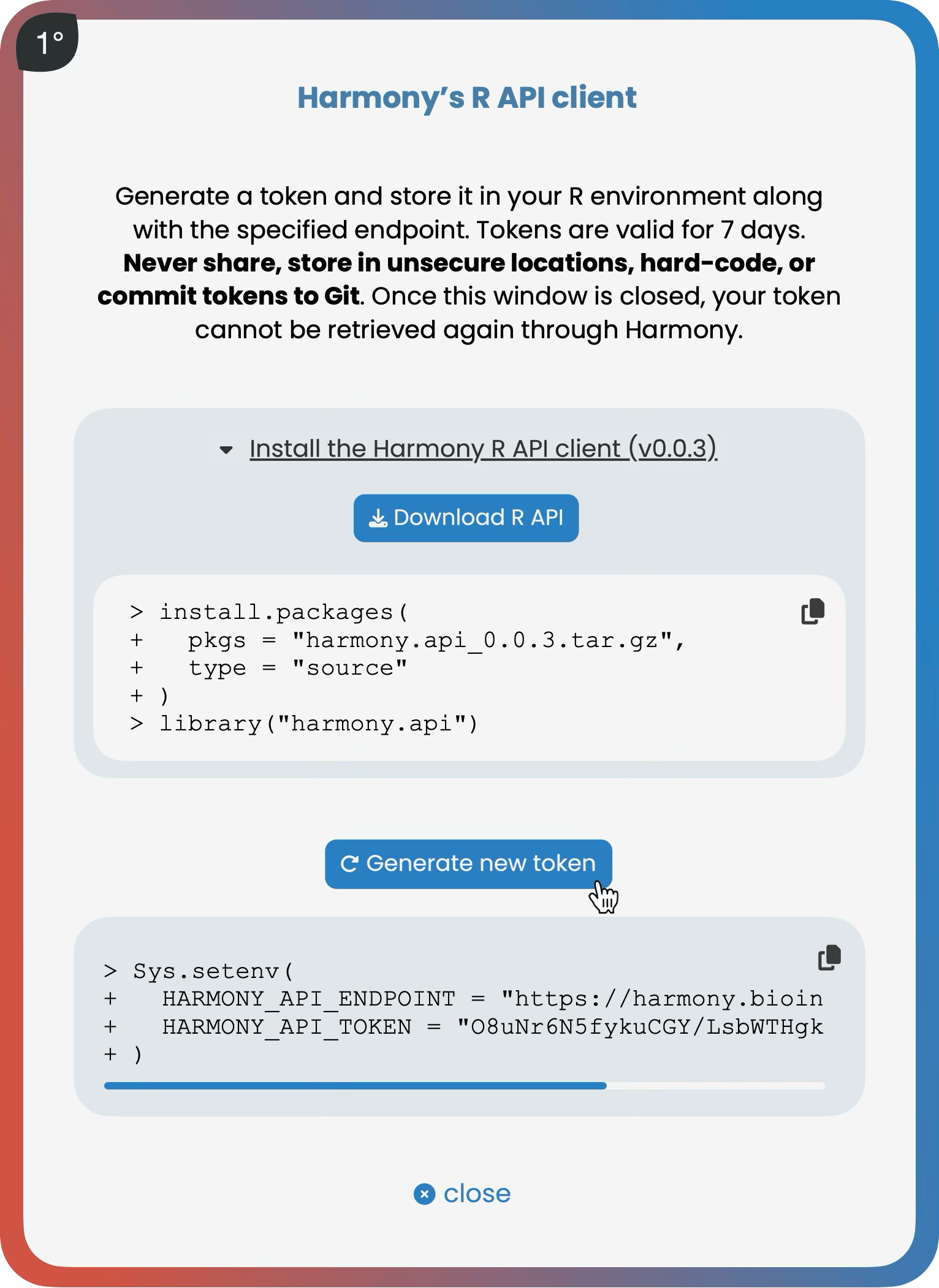
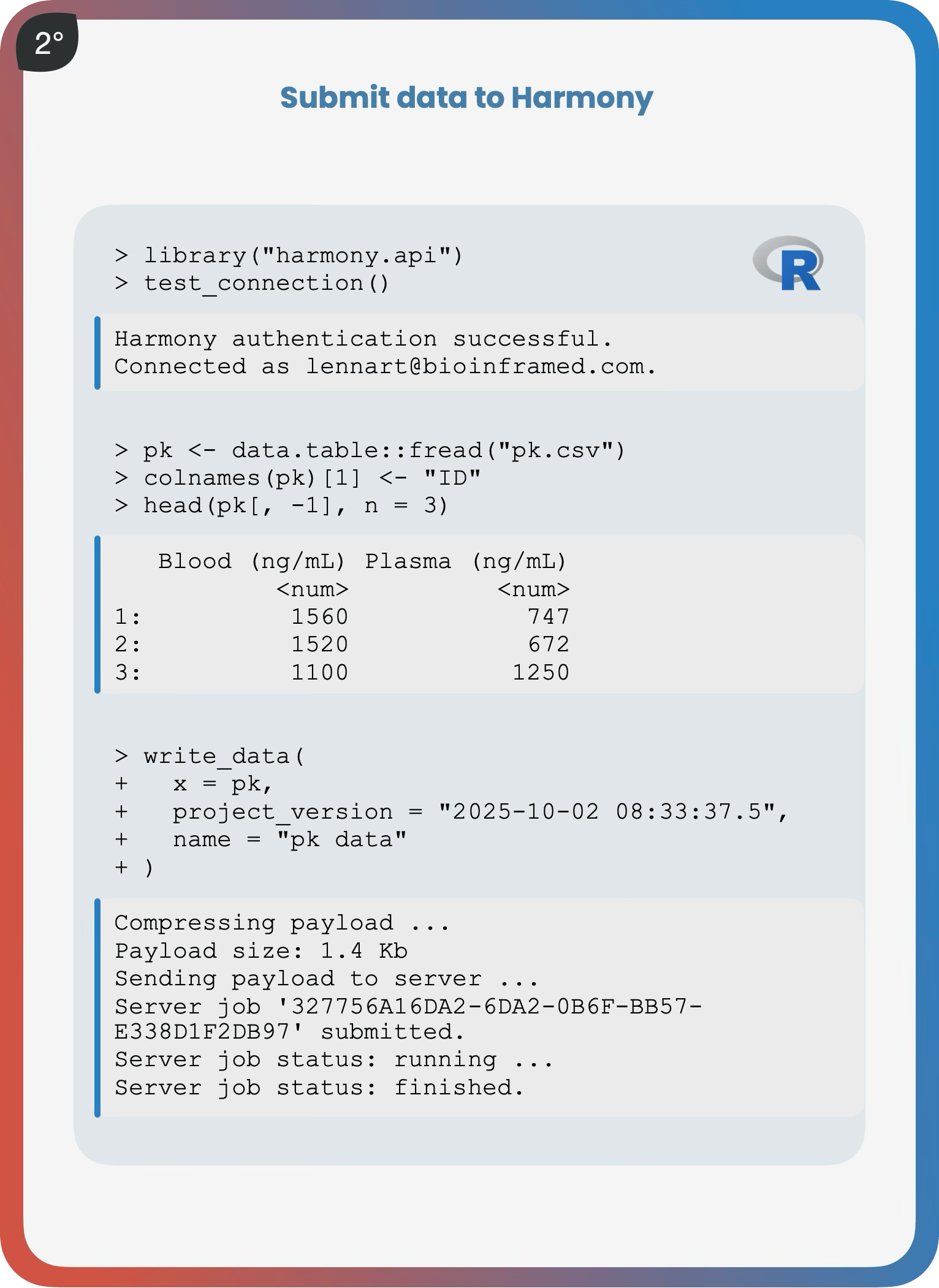
The Harmony R API
Six functions. That's the Harmony R REST API client.
1° add_data(), 2° add_metadata(), 3° write_data(), 4° write_metadata(), 5° read_data(), 6° read_metadata().
With just these six, you can assemble data from any discovery, preclinical or clinical study—whether in-house generated, CRO-delivered, downloaded from a public resource, or sequencing provider–supplied—directly and securely via HTTPS into Harmony’s database, ready for your whole team to explore interactively.
➔ Swipe ➔
Subscription license
Experience Harmony through a one-month trial license, or unlock its full capabilities with a full yearly subscription.
Trial
Duration
Users *
Data type support
Object-over-variable
Survival data
Bulk RNA seq
Single-cell data
Spatial data
Blob data
R API for coders
Frequent updates **
Feature requests **
Training sessions ***
Data curation support ***
1 month
≤ 2
✔︎
✔︎
✔︎
✘
✘
✔︎
✔︎
✘
✘
1 session
1 dataset
Full
Duration
Users *
Data type support
Object-over-variable
Survival data
Bulk RNA seq
Single-cell data
Spatial data
Blob data
R API for coders
Frequent updates **
Feature requests **
Training sessions ***
Data curation support ***
subscription
unlimited
✔︎
✔︎
✔︎
✔︎
✔︎
✔︎
✔︎
✔︎
on request
on request
on request
For detailed pricing information, to schedule a demo, or to learn about our security measures and data backup policies, let’s get in touch!
* Harmony is deployed individually for each customer on AWS infrastructure. Instance types are carefully selected based on user requirements to balance performance and cost, ensuring a smooth and responsive user experience at all times.
** The full Harmony subscription comes with frequent updates that continuously enhances usability and introduces new features. Feature requests are welcome and, if broadly applicable, are considered for implementation. Thanks to Harmony’s modular design, tailored plugins can also be developed under separate contracts to address specific needs.
*** The full Harmony subscription includes a fixed number of consultancy days that can be used for training sessions or tailored support such as data curation. Additional consultancy time is available through a separate contract if needed.